The past few decades have seen growing interest in, and understanding of, the human genome
Fueled by significant technological advances in genome sequencing, artificial intelligence and machine learning capabilities, the scientific community continues to explore this relatively new field, anticipating outcomes such as enhanced clinical applications, tailored treatments and overall improvements in mortality and healthy longevity. Initially our understanding of certain diseases, and more recently tailoring personalised treatments, was focused on single high-penetrance genetic mutations. But many more commonly encountered diseases have a polygenic architecture meaning that multiple genetic variants in combination affect disease risk. Genome-wide association studies (GWASs) have certainly helped to highlight this, and together with advances in statistical genetics, polygenic risk profiling by way of polygenic risk scores have revealed a potentially powerful tool to identify individuals at higher risk of developing certain morbidities. The wider implications of recent advances in genetics and genomics, including polygenic risk scores, need to be considered in the context of the protection industry.
The Current Landscape
The use of genetic information in insurance is understandably a highly emotive subject because of the sense of determinism and, as such, the topic usually draws a varied set of views across stakeholders. It certainly raises a number of ethical and privacy concerns so we should expect increasing scrutiny from regulators and new regulatory policies, both within and beyond the industry, to control genetic testing, any application of genetic testing, and genetic discrimination around the world. Consumer protection is utmost and we must recognise the right for anyone to have access to optimal health care or to participate in clinical trials, without fear of prejudice.
Around the world, various regulations have sought to limit the use of genetic information in insurance. In May 2017, the Canadian parliament passed Bill S-201 that completely banned genetic discrimination in insurance and the underwriting process. GINA, the Genetic Information Non‐Discrimination Act of 2008, is a federal law in the United States that protects people from genetic discrimination in health insurance and employment. Life, long term care and disability income insurance are currently exempt as lawmakers recognised that underwriting practices differ vastly from that of health insurance. Despite it not being legally binding on its member countries, the Council of Europe (COE) recommended that no genetic tests should be required for insurance purposes. Most recently, the Financial Services Council in Australia announced a moratorium in regard to genetic test results to commence from 1 July 2019. Consumers will be able to access up to AUS$500,000 lump sum cover for death and total and permanent disability, AUS$200,000 for trauma (critical illness) and AUS$4000 a month for income protection, without having to disclose adverse genetic test results.
Equally noteworthy is the rapidly falling cost of genetic testing as well as the ease of access to testing through the tremendous growth in the direct-to-consumer (DTC) genetic testing market.
In 2017 alone, over seven million DTC genetic tests were sold. The key driver was ancestry testing. People who have undergone these tests have access to significant volumes of data about their genotype. There are now applications that provide interpretive services for people who are willing to share their data. Once the data is uploaded, the app provides information on the risk of a number of different diseases and traits. The potential for information asymmetry and possible anti-selection, therefore, may need to be considered.
Genetic testing already plays a key role in clinical medicine and there are many different types of genetic tests that are currently available. This is certainly the current paradigm in oncology, but also in pre-natal and newborn screening as well as in the rare disease elucidation. There is no doubt that genomics will drive a number of additional clinical applications in the near future and, as a result, we are likely to expect both mortality and healthy longevity improvements.
A key concern for our industry is to understand the economic impact that advances in genetics and genomics will have. If regulations continue to limit the use of genetic information then the impact is clearly dependent on how useful genetic information is to risk selection. Thus, a critical question to ask is: how accurately can genomics help predict the risk of developing common diseases?
From SNP’s to GWAS to PRS to clinical application
One of the most considerable advances in the genomics space at the current time is the demonstration of polygenic risk profiling to identify disease risk. This is significant because most common diseases are not caused by a single mutation or variation in a gene (which is more straightforward to explain and predict), but result from multiple genetic variants and their interaction with lifestyle as well as environmental factors.
What is polygenic risk profiling?
The basic building blocks of the deoxyribonucleic acid (DNA) in all our cells consist of four nucleotides or base pairs. Altogether there are three billion of them that make up the complete blue print of our genetic code, and it is the order of these nucleotides that program the production of proteins and overall cell function.
Genetic variation describes the natural genetic differences that occur between individuals. Single nucleotide polymorphisms (SNPs) are individual variations in a nucleotide, where a nucleotide in a particular position in the genome is different from a reference nucleotide. A SNP is the most common type of genetic variation that exists, and we each carry millions of them, some of which we know make a difference and some which do not appear to have an impact.
Geneticists have long noticed that certain SNPs occur more commonly in people with a particular disease or trait than those without. A genome-wide association study (GWAS) is a study technique that compares those with a disease to those without a certain disease using a DNA chip or microarray. The microarray can differ in number of genetic variants studied, and usually statistical geneticists use a very low probability value to suggest a SNP is statistically significant, indicating that that a particular SNP or variation is likely to be associated with the disease or trait in question.
Genome-wide association studies have been highly successful at identifying genetic variants associated with disease. The first GWAS, conducted in 20051, compared 96 patients with age-related macular degeneration with 50 healthy controls. It identified two SNPs with significantly different allele frequency between the two study groups. An allele is one of two or more alternative forms of a gene at the same position in a chromosome.
Since this first GWAS, sample sizes have expanded, which allows SNPs with smaller odds ratios for a particular disease outcome, often at a lower frequency, to be identified. The importance of this, as mentioned, is because many common diseases are associated with multiple genetic variants that cumulatively affect disease risk.
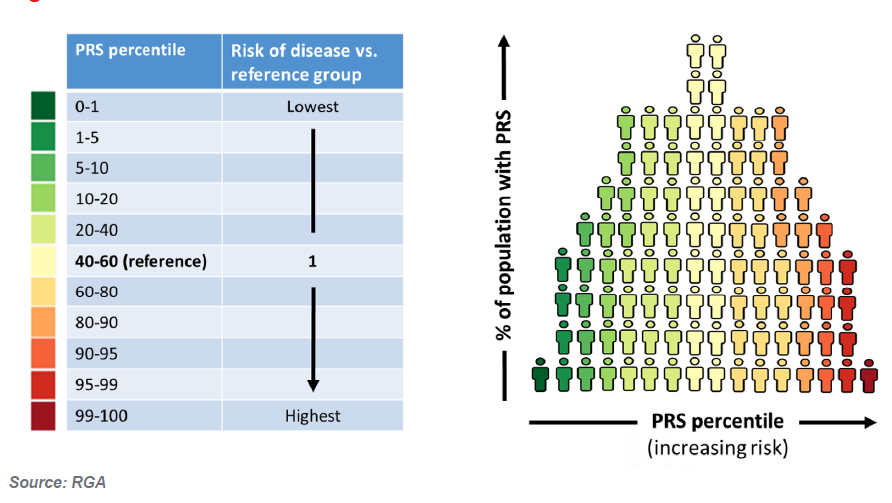
Statistical geneticists have developed the ‘polygenic risk score’ (PRS), identifying hundreds, thousands and even millions of SNPs (variants) that can be included in a single score that measures the individual’s genetic predisposition to specific diseases or traits. It is essentially an index derived by adding up the number of variants multiplied by their effect size to give an overall risk of developing a disease. PRSs are often expressed as a risk percentile. In individuals with a PRS close to the population mean, the person’s predicted genetic risk will be similar to the population’s risk, but a person with a PRS in either the 91st to 100th or 10th percentile of a population, would be considered to have the highest and lowest genetic risk, respectively. (Figure 1)
FIGURE 1:PRS DISTRIBUTION
Researchers continue to develop PRSS for many different conditions (2, 3, 4) and Table 1. Many show that there is powerful disease risk differentiation when comparing those with the highest and lowest quintiles of genetic risk.
Table 1:
Disorder | No. of Genetic Variants | Relative risk, comparing top 20% to bottom 20% PRS | Reference |
|---|
Coronary artery disease | 50 | 2.0 | Khera AV. et al. (2016), N Engl J Med. |
|---|
Coronary artery disease | 49,310 | 1.8 to 4.5 | Abraham G. et al. (2016), Eur Heart J. |
|---|
Type 2 diabetes | 1000 | 3.5 | Läll K. et al. (2017), Genet Med. |
|---|
Ischemic stroke | 10 | 1.2 to 2.0 | Hachiya T. et al. (2017), Stroke |
|---|
Breast cancer | 77 | 3.0 | Mavaddat N. et al. (2015), J Natl Cancer Inst. |
|---|
Breast cancer (East Asian ancestry) | 44 | 2.9 | Wen W. et al. (2016), Breast Cancer Res. |
|---|
Prostate cancer | 25 | 3.7 (25%) | Amin Al Olama A. et al. (2015), Cancer Epidemiol Biomarkers Prev. |
|---|
Lung cancer | 38 | 4.6 (25%) | Cheng Y. et al. (2016), Oncotarget |
|---|
Most recently a study by Khera and colleagues5, based on UK Biobank data, highlighted incredible risk stratification coming from polygenic risk scores. People with the highest 20% of PRSs for coronary artery disease, atrial fibrillation, Type II diabetes, inflammatory bowel disease and breast cancer were shown to have more than a twofold risk of each disease compared to people with a PRS in the lowest 80%. Four out of the five PRSs included almost seven million SNPs.
Equally incredible was the media attention this particular study received the day after its publication. Awareness of the potential of genetic information will undoubtedly increase as public access and attentiveness grows.
Genes, however, are not fate. Another study by Khera and colleagues6 showed that even if a person is shown to have high genetic risk for adverse cardiovascular outcomes, by living a healthy lifestyle one can mitigate this to lower their absolute risk of these events if they live healthier. Identifying those at high genetic risk could be useful for targeted public health campaigns and lifestyle interventions; the potential benefits of these and other positive effects needs also to be taken into consideration in the debate on genetics and insurance.
Indeed there is tremendous potential for the use of PRS in clinical medicine. A recent study by Abraham and colleagues4 developed a PRS to predict coronary artery disease (CAD) risk and demonstrated that people with a PRS in the highest 5% have a threefold increased risk of experiencing the condition compared to individuals with lower PRSs. They validated their PRS in two separate independent cohorts, FINRISK and Framingham Heart Study, with a combined number of 16,802 participants and 1,344 incident CAD events. After controlling for clinical risk factors including family history, the PRS still proved to be a very powerful differentiator of CAD risk. Those in the highest quintile of genetic risk compared to those in the lowest quintile of genetic risk in the Framingham cohort showed a 12 year age differential in 10% of each group going on to suffer an MI. The difference in the FINRISK cohort was even greater at 18 years.
Another potential clinical application for PRSs is in identifying those with higher cancer risk, which could lead to personalised screening programs. In a study by Mavadatt and colleagues3 looking at genetic risk and breast cancer, women whose PRSs were in the top 20% were shown to have a higher lifetime incidence of breast cancer compared with women in the lowest quintile (17.2% vs. 5.3%). Accordingly earlier screening intervention could be recommended in those women with higher genetic predisposition; these high risk women could develop breast cancer well before the usual age for population screening.
Research Collaboration
In collaboration with King’s College London13, RGA has recently looked to establish how accurately mortality and certain morbidity risk can be predicted based on detailed phenotypic information and whether such estimations could be improved by including genetic data in the form of polygenic risk scores.
This research collaboration has explored the UK Biobank data, which is a population-based cohort prospective linkage study of 500,000 participants across the UK. This incredible cohort study offers researchers an opportunity to research mortality and morbidity outcomes using genetic and environmental risk factors. In addition to typical biometric data on the participants, 20 million genetic variants have been collected.
In the RGA and King’s College London analysis, the 500 000 participants were separated into two groups, a standard and a substandard group, based on various health disclosures.
Risk prediction models were then built to study two outcomes: breast cancer and myocardial infarction incidence. Models were fully adjusted for clinical, lifestyle and socio-economic risk factors. The results demonstrated that there was roughly a two times increased risk for those in the top five percent of genetic risk compared to those in the middle percent of risk (40th to 60th percentiles) for the disease outcomes studied in both the standard group and the substandard group. This suggests that genetic predisposition significantly contributes to risk prediction above and beyond what we may include as typical underwriting risk factors.
Possible Implications and Anti-selection considerations
Sustainable life insurance availability depends on an insurer’s ability to accurately assess mortality and morbidity risk and assign a premium that aligns with the estimated risk. The growth in DTC genetic testing and the increasing ability to be able to predict disease risk through scientific advances allows consumers to access information about their disease risk that, if non-disclosed or disallowed, may be unavailable to life insurers at the time of risk assessment. This could signal an imbalance in the availability of information between life insurers and consumers.
Several research papers both outside and within the industry7-11 have looked at anti-selection risk. However these studies suggested a wide range of impact as the assumptions used in the models varied quite significantly. Furthermore, actuarial models have generally focused on high-penetrance genetic mutations. Albeit that polygenic risk profiling is still an emerging science, PRSs could play an important role in predicting common multifactorial diseases, which would greatly affect some of the genetic risk assumptions in these actuarial studies. Additionally, a challenging aspect of the modelling is the insurance risk assumptions; namely the insurance purchasing behaviour before and after performing the tests, as well as the assumptions around the impact of any risk mitigating behavior. For example, screening or other preventative lifestyle actions undertaken by the life assured once they are aware of a particular elevated disease risk. We need to acknowledge that genomic medicine may empower individuals to take preventive measures to reduce risks of future diseases, although current literature on the importance of this is conflicting12.
In Conclusion
Overall, polygenic risk scores should be considered an emerging risk issue for the protection insurance industry. Medical science will continue to push the boundaries of genetic discovery which could translate into greater predictive ability, but there is still significant work to be done. One drawback of many of the current published scientific research is that its application is limited to Caucasian populations and so more evidence from non-Caucasian cohorts needs to emerge for wider adoption and applicability. Education for clinicians and the public is crucial as we all begin to navigate this new world. Industry supervisory bodies will continue to monitor and regulate this space. This is a time for both risk and opportunity in the industry. Undoubtedly, polygenic risk scores will be a specific area of focus as we continue to explore this fascinating field of scientific discovery, particularly as we seek to better understand common disease risk and outcomes through our genome.